Uncanny Resemblances: When a Heating Coil is like an Adaptive Peak and a Golden Retriever is like Random Genetic Drift
Some of the most interesting questions in evolutionary biology arise from both unexpected diversity and unexpected similarity among organisms. Sometimes organisms are genetically similar, yet display outward characteristics (phenotypes) that can be strikingly different. Other times, organisms display phenotypes that are astonishingly similar despite greater genetic distances. As Kristen explained in our previous post, factors such as trait covariance (bounded by genetic, developmental, and integrative limitations) can have a notable effect on the frequency of observed phenotypes in a population. How those limitations act together to restrict the possible directions a phenotype can evolve defines something we refer to as a morphospace.
Morphospaces
We describe a morphospace using many different phenotypes simultaneously. If we can characterize a sufficient subset of the conditions (parameters) that bound a given morphospace, we might be able to learn something about how different evolutionary forces affect the distribution of phenotypes within that morphospace. As we increase the number of phenotypes that we are considering, though, we increase the number of dimensions needed to describe a given morphospace.
While several multivariate methods exist to deal with statistical comparisons of such high-dimension datasets, consensus on which method is most reliable has yet to be reached. This has made direct, meaningful comparisons of populations in multivariate morphospace difficult to achieve. Notwithstanding, if we can begin to uncover how those factors both limit and allow phenotypic differences to arise not just within one species, but between multiple closely related species, we can explore novel questions. For example, will two closely related species converge on similar phenotypes (convergent evolution) or diverge into dissimilar phenotypes (divergent evolution) when the given species approach natural selection fields?
Natural selection fields?
In the case of convergence in two closely related species, the phenotype in question could have arisen in each species by chance (Brownian motion) or the phenotype may have arisen in each species in response to a similar instance of natural selection (an adaptive peak). As Kristen previously pointed out, we calculate the strength of natural selection, using a variant of Russel Lande’s equations, as a vector of gradients that shift a given mean phenotype z at some time t to a new value zꞌ at time tꞌ: Δz = Pβ (Lande 1979; Lande and Arnold 1983). A gradient can be interpreted as the representation of a dynamic field, such as temperature in a room. When the room is undisturbed, we can assume that the temperature is statistically uniform throughout. However, if we were to place a heating coil in the center of the room, the distribution of temperatures about the room would change. At the edges of the room, temperature might increase slightly, but as we move towards the coil, the temperature as well as the rate of temperature change itself will increase. This increase produces a temperature peak (local maximum) in the temperature field in that room.
If two objects with like properties approach that peak, we can expect the objects to experience a parallel change in temperature. By extension, we can imagine natural selection as a field in its own right. When there is little to no natural selection, the field looks much like the temperature distribution in our undisturbed room, statistically homogeneous. When an instance of natural selection arises in the field, we get a peak. The peak can be described as a selective maximum (with respect to the intensity of natural selection) or as an adaptive peak (with respect to the effect of selection on a given phenotype or group of phenotypes). Using the second description, Δz can be understood as the shift from a sub-optimal phenotype (one that is being selected against) towards an optimal phenotype (one that is selected for) in cases where β is statistically significant.
Leveraging Adaptive Peaks
If we can demonstrate that a phenotype has been shaped by a hypothesized adaptive peak in a population, can we make any assumptions about a similar or identical phenotype in a closely related population? Specifically, when we observe a convergent phenotype in two closely related species, is there some way to ascertain whether these two phenotypes arose in response to the same (or similar) instance of natural selection? The answer to this question is paramount when trying to resolve complex phylogenies. A mathematical model proposed by Leonard Ornstein and George Uhlenbeck, later elaborated by Thomas Hansen, and finally placed within a phylogenetic modeling framework by Marguerite Butler and Aaron King, helps to address this question (Butler and King 2004; Cressler et al. 2015; Hansen 1997; Uhlenbeck and Ornstein 1930). The following equation describes how and what we are modeling:
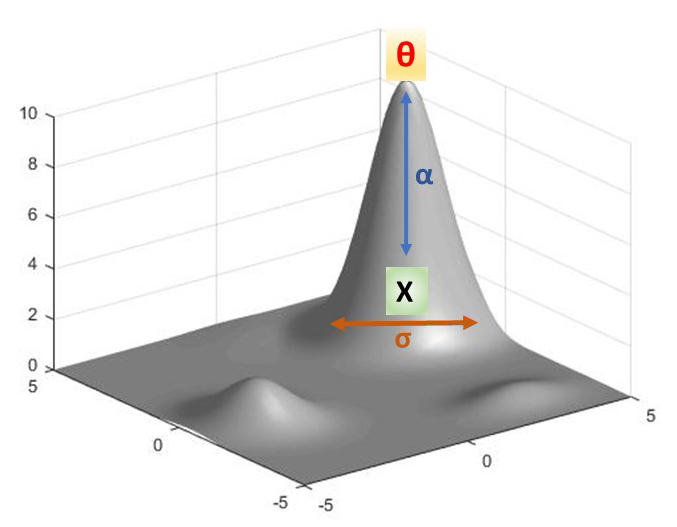
dX(t) = α[θ - X(t)]dt + σdB(t)
where α is the rate of approach of a given phenotype, X, toward the adaptive peak, θ (conversely the strength of natural selection’s pull from the sub-optimal to the optimal state), and σ is the rate of motion of a given phenotype towards a random step whose location is randomly determined by Β. If we imagine an adventurous golden retriever on a leash chasing a squirrel, α can be understood as the strength of a tug on the leash by the frustrated owner while σ is the pull on the leash due to the frantic wandering of our golden retriever back and forth in a direction orthogonal (at right angles) to the direction of the owner’s tug at time t. The term θ - X(t) then is the position of the dog (X) relative to where the owner is standing (θ) at some time t while the term dB(t) describes the location of the random orthogonal position of the dog at that same time t. This equation then allows us to simultaneously model the effect of natural selection as well as stochasticity (randomness or noise) on phenotypic evolution over time.
Applying Theory to Real Biological Systems
Macaca arctoides, courtesy http://www.nzmc.org/ztg/491
Macaca radiata, courtesy https://birdsphotos.wordpress.com/animals/mammal/
Having been initially trained as a paleoanthropologist, my instinct is to use this theoretical framework to investigate human phenotypic variation. However, as we are the only extant species of our genus, we must instead turn to other primate groups to model phenotypic change between closely related species. For that reason, my current work instead focuses on another Old World primate group, the genus Macaca (macaque monkeys). Much like our Homo ancestors, populations of macaques migrated out of Africa, and ultimately spread across Eurasia spanning multiple latitudes, altitudes, and ecological zones. They are the most dispersed primate group apart from humans, and thus provide an excellent opportunity to understand the interplay between genetics, environment, and form that have shaped the distribution of macaque phenotypes we observe today.
My pilot research (Williams and Auerbach, 2016) supported directional selection acting on facial traits of the species M. radiata, which inhabits colder regions, supporting prior research into ecogeographical patterning among them (Fooden 1982; Ito et al. 2014; Weinstein 2011). That same pilot study found support for directional selection acting on the face of two other species, M. fascicularis and M. nemestrina as well, although clearly not in the same way as M. radiata. My current project leverages Orenstein and Uhlenbeck ‘s work to address two questions left unanswered by my pilot study: 1) Are there other macaques that share facial phenotypes with these three species? 2) Are there selective pressures beyond those factors associated with ecogeography, such as locomotor demands, foraging mechanics, or shared population history, that better account for the patterns of convergence?
My preliminary results show there are additional macaque species sharing similar facial traits, supporting convergence between three pairs of macaques: M. radiata and M. arctoides, M. nemestrina and M. silenus, and M. mulatta and M. fascicularis. However, this convergence is not uniform across phenotypes, so the second question remains open. There is, though, good support for ecogeographic factors underlying the convergence in facial phenotypes between M. radiata and M. arctoides. I am currently expanding the number of macaque species in the analysis, which along with new additions to the phenotypes I have already been measuring will allow me to test hypotheses that address locomotion, foraging, and population history. I will be presenting the results of this work in New Orleans at the Annual Meeting of the American Association of Physical Anthropologists in April and will share them here with you as well, so stay tuned!
References
Fooden J. 1982. Ecogeographic segregation of macaque species. Primates 23(4):574-579.